
VERA PANCALDI
ASSOCIATE INVESTIGATORS: FLAVIEN RAYNAL, MIGUEL madrid & hadi kabalane
Epigenomics And Chromatin Networks
Chromosome Conformation Capture (3C) techniques demonstrate that the organization of chromatin in the cell nucleus is a fundamental aspect to consider in order to understand the regulation of cellular phenotypes. Our team uses biological networks to represent chromatin and understand how different epigenetic factors impact cellular phenotypes (differentiation, cancer proliferation, immune cell status), which may play an important role in tumor progression and response to immune therapies.
We have recently developed 2 tools for the scientific community interested in integrating epigenomic data in a chromatin structure context: GARDEN-NET is a 3D genome browser accessible via a web interface, ChAseR is an R package facilitating the integration epigenomic datasets with chromatin contact networks and calculation of assortativity, which estimates an association between a given feature (also user defined) and 3D interactions.
One of the main challenges remains the understanding of the relationship between plasticity and variability of cellular and epigenetic phenotypes.
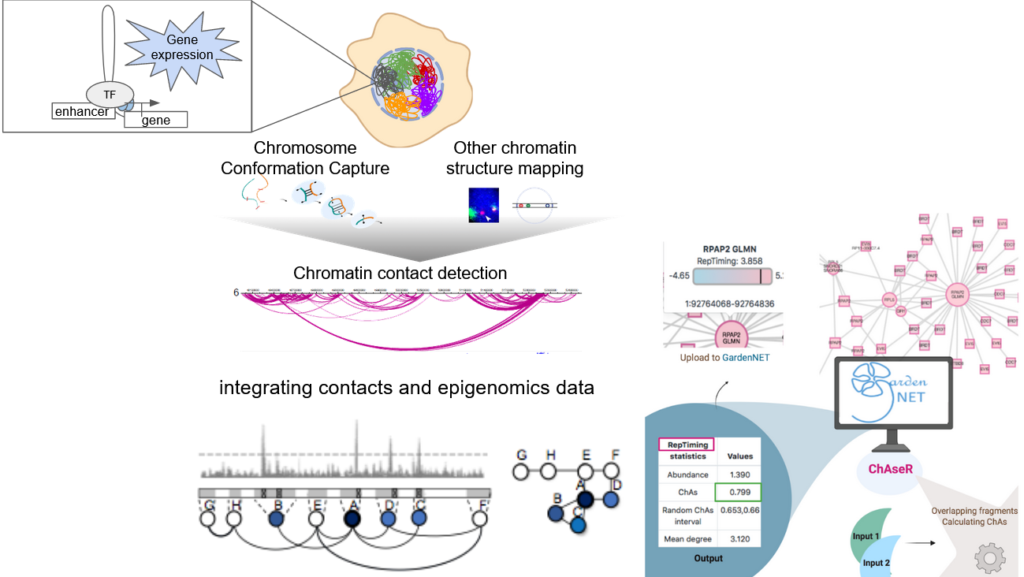
The relationship between chromatin organization within the nucleus and gene expression is still unclear. We have developed methods to integrate datasets that describe the structure of the genome in terms of 3D interactions with other datasets using network theory. We represent the genome as a network and apply network theory concepts to study the principles of 3D genome organization.
Madrid-Mencia et al. 2020 https://academic.oup.com/nar/article/48/8/4066/5809158?login=true
Github https://github.com/VeraPancaldiLab/GARDEN-NET
https://github.com/VeraPancaldiLab/ChAseR_demo
Pancaldi 2021 https://www.frontiersin.org/articles/10.3389/fbinf.2021.742216/full
CRCT Chair of Informatics in Oncology (Fondation Toulouse Cancer Sante, Inserm et Pierre Fabre)

