team
VERA PANCALDI
NetB(IO)² :
Network Biology for Immuno-oncology
THE SPECIFITIES
OF OUR RESEARCH AXIS
Our lab studies the relationship between patients and cancer, focusing on how the immune system and the tumour interact. We aim to improve therapies that exploit this interaction. Currently such treatments only work on some patients and, often, for a limited time before they develop resistance against the therapy.
Our goal is to understand how variability in the patients, in the tumours, and in their interactions affects the efficacy of these therapies. We use computational approaches including data science, mathematical modeling and especially network theory, as well as several biological model systems.
Networks are a useful framework to represent systems in which relationships such as interactions or similarity between objects are important. All networks share some common properties and a vast literature exists on how to measure them. The beauty is that our understanding of one type of network can help us better understand a completely different one.
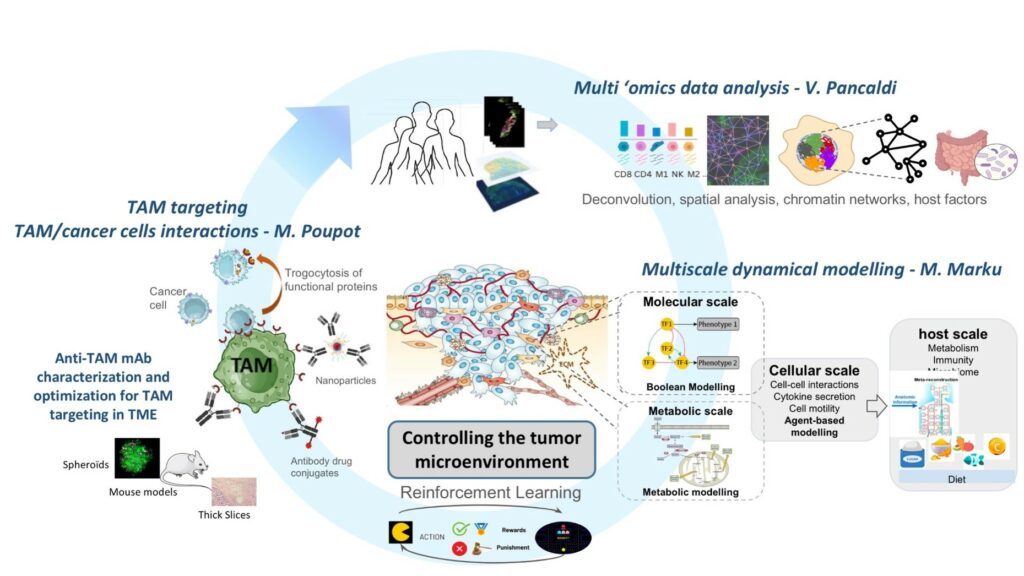
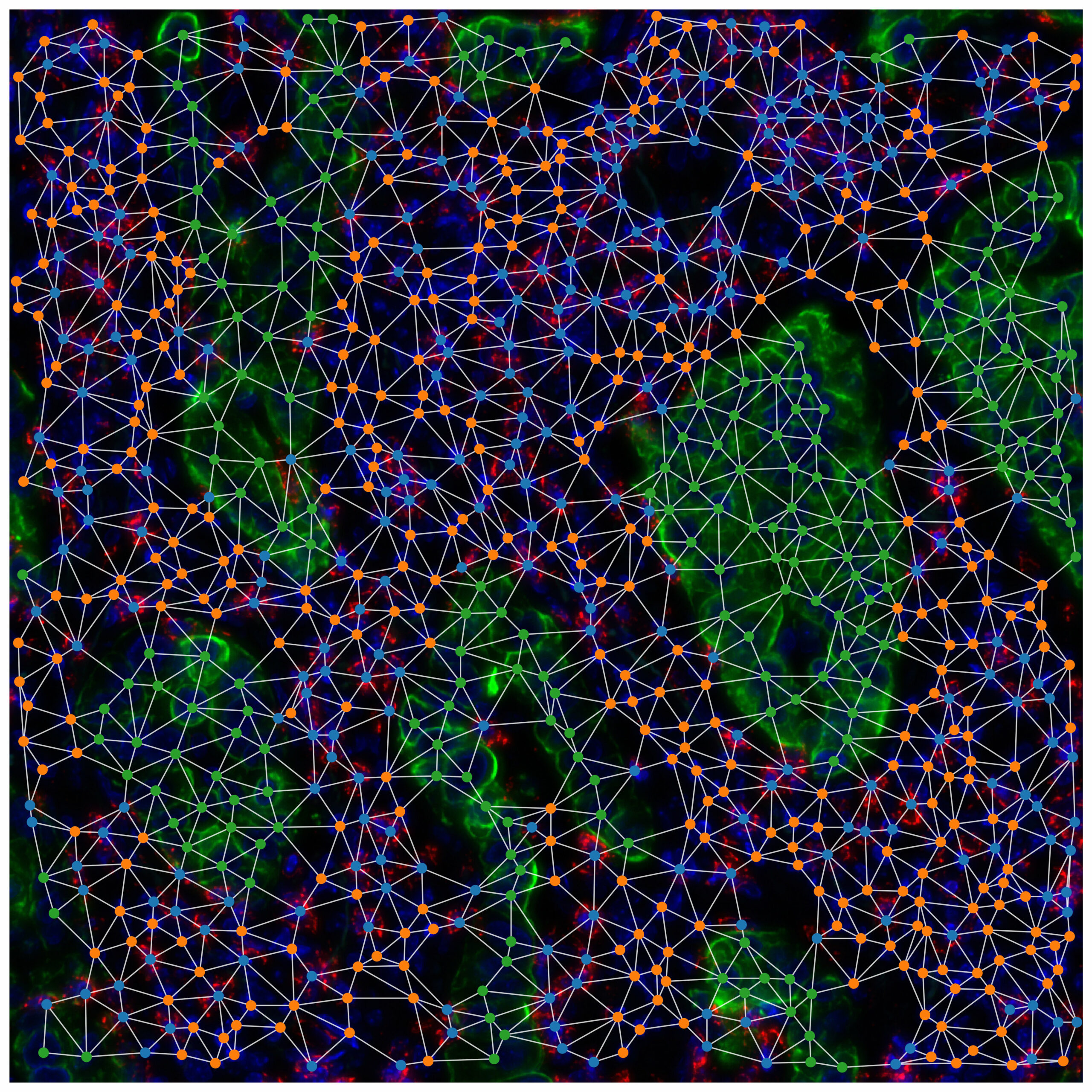
We apply network models to study such diverse systems as networks of patient-patient similarity, 3D interaction networks of genes in the nucleus and networks of interactions between different cell types in a tumour.
There are many contexts within the body that are characterised by a heterogeneous mix of cell types. In tissues, complex relationships between cell types establish ‘ecological networks’ whose properties reflect the equilibria that define health against disease. In tumours, cancer cells are surrounded by extremely heterogeneous cell populations, including stromal and immune cells, whose spatial patterns and specific interactions strongly impact response to therapies and patient outcomes.
Simulating the complex interactions of these cells in our computers, both at the molecular and intercellular level, offers the possibility to study these systems in a multitude of scenarios. This is an excellent way to formulate new hypotheses and allows better use of our experimental resources.
Our main research objectives are the following
1) Characterising cell populations and their interactions in the tumor microenvironment (TME) considering cell-cell communication networks and spatial patterns as extracted from transcriptomics (both bulk and single-cell) or spatial omics and imaging produced from patient samples as well as their epigenomic landscape.
2) Simulating dynamic cell processes and inter-cellular interactions in the TME involving cancer, immune and stromal cells, exploiting mathematical models in different scales (molecular, cellular, etc.) of in vitro (2D/3D) and in vivo cancer models.
3) Developing strategies to control the tumour microenvironment to improve sensitivity of cancer cells to treatments exploiting an in-house patented monoclonal antibody specifically targeting tumor associated macrophages, identifying new strategies for TME reprogramming, and expanding our point of view to patient characteristics (immune fitness, microbiome, exposures, etc.)
Computational Biology
Network Theory
Heterogeneity and Tumour Microenvironment
Immune System
Epigenomics
Spatial and single cell transcriptomics
Genome Architecture
Mathematical Models
research projects
THE TEAM’S
FOCUS
Discover
Understand
Live
Just a few hours to go for early bird registration for our @netbiomed2025 symposium, join us in Maastricht on June 2nd. Check out our website for registration instructions #AI #DigitalTwins @net_science @NetSciConf
3 days before @NetSci2025 early bird ends: Don't miss your chance to attend @netbiomed2025. Please check out our website for details 7 Amazing speakers https://netbiomed2025.github.io AI Digital twins @BSC_CNS @CNIOStopCancer @esantapau @CamNetNet @net_science @ConfCompSys @SysBioCurie
SCIENTIFIC PRODUCTIONS
PUBLICATIONS 2025
PUBLICATIONS 2024
PUBLICATIONS 2023
PUBLICATIONS 2022
PUBLICATIONS 2021
PUBLICATIONS 2020
PUBLICATIONS 2019
TEAM MEMBERS
PARTNERSHIPS & funding

Centre de Recherches en Cancérologie de Toulouse (Oncopole)
Toulouse – FR
CRCT-Oncopole Follow 1,433 1,279
Compte officiel du Centre de Recherches en Cancérologie de Toulouse
🇬🇧Cytidine deaminase-dependent mitochondrial biogenesis as a potential vulnerability in pancreatic cancer cells. ImPact team @CordelierPierre article from @crctoncopole published in @CommsBio
➡️https://www.crct-inserm.fr/en/targeting-the-energy-productio...
Nous contacter
05 82 74 15 75
Envie de rejoindre
L’équipe du CRCT ?